Train Graph Neural Networks with AUPRC Loss
Introduction
In this tutorial, you will learn how to quickly train a GNN
model by optimizing AUPRC with our novel APLoss and SOAP optimizer
[ref] on a binary molecule
classification task from ogbg-molpcba dataset. After completion of this
tutorial, you should be able to use LibAUC to train your own models
on your own datasets.
Reference:
If you find this tutorial helpful, please acknowledge our library and cite the following papers:
@inproceedings{yuan2023libauc,
title={LibAUC: A Deep Learning Library for X-Risk Optimization},
author={Zhuoning Yuan and Dixian Zhu and Zi-Hao Qiu and Gang Li and Xuanhui Wang and Tianbao Yang},
booktitle={29th SIGKDD Conference on Knowledge Discovery and Data Mining},
year={2023}
}
@article{qi2021stochastic,
title={Stochastic Optimization of Areas Under Precision-Recall Curves with Provable Convergence},
author={Qi, Qi and Luo, Youzhi and Xu, Zhao and Ji, Shuiwang and Yang, Tianbao},
journal={Advances in Neural Information Processing Systems},
volume={34},
year={2021}
}
Install LibAUC
Let’s start with installing our library here. In this tutorial, we will use the lastest version for LibAUC by using pip install -U.
!pip install -U libauc
Importing LibAUC
Import required packages to use
from libauc.losses import APLoss
from libauc.optimizers import SOAP
from libauc.models import GCN, DeeperGCN, GIN, GINE, GAT, MPNN, GraphSAGE, PNA
from libauc.sampler import DualSampler
from libauc.metrics import auc_prc_score
from ogb.graphproppred import PygGraphPropPredDataset
import torch
import torch_geometric
import numpy as np
import pandas as pd
import os
Reproducibility
The following function set_all_seeds limits the number of sources
of randomness behaviors, such as model intialization, data shuffling,
etcs. However, completely reproducible results are not guaranteed
across PyTorch releases
[Ref].
def set_all_seeds(SEED):
# REPRODUCIBILITY
np.random.seed(SEED)
torch.manual_seed(SEED)
torch.cuda.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
set_all_seeds(2023)
Loading datasets
Now, we define the data input pipeline. We inherit the PygGraphPropPredDataset class from ogb package since we are going to work on ogbg-molpcba dataset
class GraphDataset(PygGraphPropPredDataset):
def __getitem__(self, idx):
if isinstance(idx, (int,np.int64)):
item = self.get(self.indices()[idx])
item.idx = torch.LongTensor([idx])
return item
else:
return self.index_select(idx)
For simplicity of exposition, we work on the first task from ogbg-molpcba dataset.
dataset = GraphDataset(name = 'ogbg-molpcba')
labels = pd.read_csv(os.path.join('./dataset/ogbg_molpcba/raw', 'graph-label.csv.gz'),
compression='gzip', header = None).values
#### get the official train_val_test split
split_idx = dataset.get_idx_split()
#### get training lable for task_0
train_labels = labels[split_idx["train"]][:,0]
#### remove samples which have 'nan' as their labels
not_nan = ~np.isnan(train_labels)
train_labels = train_labels[not_nan]
train_dataset = dataset[split_idx["train"]][not_nan]
test_dataset = dataset[split_idx["test"]][~np.isnan(labels[split_idx["test"]][:,0] )]
Configuration
Hyper-Parameters
# Hyper-Parameters
batch_size = 64
lr = 1e-3
margin = 0.6
gamma = 0.1
weight_decay = 1e-5
total_epoch = 60
decay_epoch = [30, 45]
load_pretrain = False
with_edge_features = True
Pretraining (Recommended)
Following the original paper, it’s recommended to start from a pretrained checkpoint with cross-entropy loss to significantly boost models’ performance. It includes a pre-training step with standard cross-entropy loss, and an AUPRC maximization step that maximizes an AUPRC surrogate loss of the pre-trained model.
from torch.optim import Adam
import warnings
warnings.filterwarnings('ignore')
load_pretrain = True
### ************ model selection BEGIN***************#############
##### without edge feature
# model = GCN(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.5, jk = "last", norm='BatchNorm')
# model = GIN(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.5, jk = "last", norm='BatchNorm')
# model = GraphSAGE(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.2, jk = "last", norm='BatchNorm')
##### with edge feature
# model = DeeperGCN(emb_dim=300, num_tasks= 1, num_layers = 14, t=0.1, learn_t=True, dropout=0.0, norm='BatchNorm')
model = GINE(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.5, jk = "last", norm='BatchNorm')
# model = GAT(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.0, jk = "last",act='elu',norm='BatchNorm',v2=False, heads=6)
# model = GAT(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.0, jk = "last",act='elu', norm='BatchNorm',v2=True, heads=6)
# model = MPNN(emb_dim=100, num_tasks= 1,num_layers = 3, dropout=0.2, jk = "last", norm='BatchNorm')
# # for PNA
# # Compute the maximum in-degree in the training data.
# from torch_geometric.utils import degree
# max_degree = -1
# for data in train_dataset:
# d = degree(data.edge_index[1], num_nodes=data.num_nodes, dtype=torch.long)
# max_degree = max(max_degree, int(d.max()))
# # Compute the in-degree histogram tensor
# deg = torch.zeros(max_degree + 1, dtype=torch.long)
# for data in train_dataset:
# d = degree(data.edge_index[1], num_nodes=data.num_nodes, dtype=torch.long)
# deg += torch.bincount(d, minlength=deg.numel())
# model = PNA(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.2, jk = "last", norm='BatchNorm', towers=5, deg=deg)
### ************ model selection END***************#############
model = model.cuda()
loss_fn = torch.nn.BCELoss()
optimizer =Adam(model.parameters(), lr=lr, weight_decay=weight_decay)
trainloader = torch_geometric.loader.DataLoader(train_dataset, batch_size=batch_size, shuffle=True, num_workers=2)
testloader = torch_geometric.loader.DataLoader(test_dataset, batch_size=batch_size, shuffle=False, num_workers=2)
best_test = 0
for epoch in range(total_epoch):
if epoch in decay_epoch:
for param_group in optimizer.param_groups:
param_group['lr'] = 0.1 * param_group['lr']
model.train()
for step, batch in enumerate(trainloader):
batch = batch.cuda()
if with_edge_features: #### for DeeperGCN, GINE, GAT, MPNN
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.edge_attr, batch.batch) )
else: #### for GCN, GIN, GraphSAGE
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.batch) )
loss = loss_fn(pred, batch.y[:,0].reshape(-1,1))
optimizer.zero_grad()
loss.backward()
optimizer.step()
######***evaluation***####
# evaluation on test sets
model.eval()
test_pred_list, test_true_list = [], []
with torch.no_grad():
for step, batch in enumerate(testloader):
batch = batch.cuda()
if with_edge_features: #### for DeeperGCN, GINE, GAT, MPNN
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.edge_attr, batch.batch) )
else: #### for GCN, GIN, GraphSAGE
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.batch) )
test_pred_list.append(pred.cpu().detach().numpy())
test_true_list.append(batch.y[:,0].cpu().detach().numpy())
test_true = np.concatenate(test_true_list)
test_pred = np.concatenate(test_pred_list)
test_ap = auc_prc_score(test_true, test_pred)
if best_test < test_ap:
best_test = test_ap
torch.save(model.state_dict(), 'ce_pretrained_model_soap_mol.pth')
model.train()
print("epoch: %s, test_ap: %.4f, best_test_ap: %.4f, lr: %.4f"%(epoch, test_ap, best_test, optimizer.param_groups[0]['lr'] ))
epoch: 0, test_ap: 0.2096, best_test_ap: 0.2096, lr: 0.0010
epoch: 1, test_ap: 0.2193, best_test_ap: 0.2193, lr: 0.0010
epoch: 2, test_ap: 0.2327, best_test_ap: 0.2327, lr: 0.0010
epoch: 3, test_ap: 0.2550, best_test_ap: 0.2550, lr: 0.0010
epoch: 4, test_ap: 0.2570, best_test_ap: 0.2570, lr: 0.0010
epoch: 5, test_ap: 0.2581, best_test_ap: 0.2581, lr: 0.0010
epoch: 6, test_ap: 0.2696, best_test_ap: 0.2696, lr: 0.0010
epoch: 7, test_ap: 0.2824, best_test_ap: 0.2824, lr: 0.0010
epoch: 8, test_ap: 0.2865, best_test_ap: 0.2865, lr: 0.0010
epoch: 9, test_ap: 0.2963, best_test_ap: 0.2963, lr: 0.0010
epoch: 10, test_ap: 0.2955, best_test_ap: 0.2963, lr: 0.0010
epoch: 11, test_ap: 0.2883, best_test_ap: 0.2963, lr: 0.0010
epoch: 12, test_ap: 0.3053, best_test_ap: 0.3053, lr: 0.0010
epoch: 13, test_ap: 0.3117, best_test_ap: 0.3117, lr: 0.0010
epoch: 14, test_ap: 0.3152, best_test_ap: 0.3152, lr: 0.0010
epoch: 15, test_ap: 0.3192, best_test_ap: 0.3192, lr: 0.0010
epoch: 16, test_ap: 0.3112, best_test_ap: 0.3192, lr: 0.0010
epoch: 17, test_ap: 0.3149, best_test_ap: 0.3192, lr: 0.0010
epoch: 18, test_ap: 0.3193, best_test_ap: 0.3193, lr: 0.0010
epoch: 19, test_ap: 0.3211, best_test_ap: 0.3211, lr: 0.0010
epoch: 20, test_ap: 0.3047, best_test_ap: 0.3211, lr: 0.0010
epoch: 21, test_ap: 0.3215, best_test_ap: 0.3215, lr: 0.0010
epoch: 22, test_ap: 0.3243, best_test_ap: 0.3243, lr: 0.0010
epoch: 23, test_ap: 0.3198, best_test_ap: 0.3243, lr: 0.0010
epoch: 24, test_ap: 0.3359, best_test_ap: 0.3359, lr: 0.0010
epoch: 25, test_ap: 0.3312, best_test_ap: 0.3359, lr: 0.0010
epoch: 26, test_ap: 0.3150, best_test_ap: 0.3359, lr: 0.0010
epoch: 27, test_ap: 0.3228, best_test_ap: 0.3359, lr: 0.0010
epoch: 28, test_ap: 0.3225, best_test_ap: 0.3359, lr: 0.0010
epoch: 29, test_ap: 0.3333, best_test_ap: 0.3359, lr: 0.0010
epoch: 30, test_ap: 0.3484, best_test_ap: 0.3484, lr: 0.0001
epoch: 31, test_ap: 0.3517, best_test_ap: 0.3517, lr: 0.0001
epoch: 32, test_ap: 0.3555, best_test_ap: 0.3555, lr: 0.0001
epoch: 33, test_ap: 0.3548, best_test_ap: 0.3555, lr: 0.0001
epoch: 34, test_ap: 0.3573, best_test_ap: 0.3573, lr: 0.0001
epoch: 35, test_ap: 0.3606, best_test_ap: 0.3606, lr: 0.0001
epoch: 36, test_ap: 0.3573, best_test_ap: 0.3606, lr: 0.0001
epoch: 37, test_ap: 0.3578, best_test_ap: 0.3606, lr: 0.0001
epoch: 38, test_ap: 0.3582, best_test_ap: 0.3606, lr: 0.0001
epoch: 39, test_ap: 0.3606, best_test_ap: 0.3606, lr: 0.0001
epoch: 40, test_ap: 0.3598, best_test_ap: 0.3606, lr: 0.0001
epoch: 41, test_ap: 0.3628, best_test_ap: 0.3628, lr: 0.0001
epoch: 42, test_ap: 0.3575, best_test_ap: 0.3628, lr: 0.0001
epoch: 43, test_ap: 0.3582, best_test_ap: 0.3628, lr: 0.0001
epoch: 44, test_ap: 0.3602, best_test_ap: 0.3628, lr: 0.0001
epoch: 45, test_ap: 0.3624, best_test_ap: 0.3628, lr: 0.0000
epoch: 46, test_ap: 0.3603, best_test_ap: 0.3628, lr: 0.0000
epoch: 47, test_ap: 0.3615, best_test_ap: 0.3628, lr: 0.0000
epoch: 48, test_ap: 0.3621, best_test_ap: 0.3628, lr: 0.0000
epoch: 49, test_ap: 0.3617, best_test_ap: 0.3628, lr: 0.0000
epoch: 50, test_ap: 0.3627, best_test_ap: 0.3628, lr: 0.0000
epoch: 51, test_ap: 0.3630, best_test_ap: 0.3630, lr: 0.0000
epoch: 52, test_ap: 0.3637, best_test_ap: 0.3637, lr: 0.0000
epoch: 53, test_ap: 0.3618, best_test_ap: 0.3637, lr: 0.0000
epoch: 54, test_ap: 0.3624, best_test_ap: 0.3637, lr: 0.0000
epoch: 55, test_ap: 0.3624, best_test_ap: 0.3637, lr: 0.0000
epoch: 56, test_ap: 0.3620, best_test_ap: 0.3637, lr: 0.0000
epoch: 57, test_ap: 0.3625, best_test_ap: 0.3637, lr: 0.0000
epoch: 58, test_ap: 0.3630, best_test_ap: 0.3637, lr: 0.0000
epoch: 59, test_ap: 0.3631, best_test_ap: 0.3637, lr: 0.0000
Optimizing AUPRC Loss
We define dataset, DualSampler and dataloader here. By
default, we use batch_size 64 and we oversample the minority
class with pos:neg=1:1 by setting sampling_rate=0.5.
sampling_rate = 0.5
sampler = DualSampler(None, batch_size, labels=train_labels, sampling_rate=sampling_rate)
trainloader = torch_geometric.loader.DataLoader(train_dataset, batch_size=batch_size, sampler=sampler, num_workers=2)
trainloader_eval = torch_geometric.loader.DataLoader(train_dataset, batch_size=batch_size, shuffle=False, num_workers=2)
testloader = torch_geometric.loader.DataLoader(test_dataset, batch_size=batch_size, shuffle=False, num_workers = 2)
Model, Loss and Optimizer
### ************ model selection BEGIN***************#############
##### without edge feature
# model = GCN(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.5, jk = "last", norm='BatchNorm')
# model = GIN(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.5, jk = "last", norm='BatchNorm')
# model = GraphSAGE(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.2, jk = "last", norm='BatchNorm')
##### with edge feature
# model = DeeperGCN(emb_dim=300, num_tasks= 1, num_layers = 14, t=0.1, learn_t=True, dropout=0.0, norm='BatchNorm')
model = GINE(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.5, jk = "last", norm='BatchNorm')
# model = GAT(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.0, jk = "last",act='elu',norm='BatchNorm',v2=False, heads=6)
# model = GAT(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.0, jk = "last",act='elu', norm='BatchNorm',v2=True, heads=6)
# model = MPNN(emb_dim=100, num_tasks= 1,num_layers = 3, dropout=0.2, jk = "last", norm='BatchNorm')
# # for PNA
# # Compute the maximum in-degree in the training data.
# from torch_geometric.utils import degree
# max_degree = -1
# for data in train_dataset:
# d = degree(data.edge_index[1], num_nodes=data.num_nodes, dtype=torch.long)
# max_degree = max(max_degree, int(d.max()))
# # Compute the in-degree histogram tensor
# deg = torch.zeros(max_degree + 1, dtype=torch.long)
# for data in train_dataset:
# d = degree(data.edge_index[1], num_nodes=data.num_nodes, dtype=torch.long)
# deg += torch.bincount(d, minlength=deg.numel())
# model = PNA(emb_dim=300, num_tasks= 1,num_layers = 5, dropout=0.2, jk = "last", norm='BatchNorm', towers=5, deg=deg)
### ************ model selection END***************#############
model = model.cuda()
# load pretrained model
if load_pretrain:
PATH = 'ce_pretrained_model_soap_mol.pth'
state_dict = torch.load(PATH)
filtered = {k:v for k,v in state_dict.items() if 'graph_pred_linear' not in k}
msg = model.load_state_dict(filtered, False)
print(msg)
model.graph_pred_linear.reset_parameters()
loss_fn = APLoss(len(train_dataset), margin=margin, gamma=gamma)
optimizer = SOAP(model.parameters(), lr=lr, mode='adam', weight_decay=weight_decay)
Training
Now it’s time for training. And we evaluate Average Precision performance after every epoch.
print ('Start Training')
print ('-'*30)
train_log, test_log = [], []
best_test = 0
for epoch in range(total_epoch):
if epoch in decay_epoch:
optimizer.update_lr(decay_factor=10)
model.train()
for step, batch in enumerate(trainloader):
batch = batch.cuda()
if with_edge_features: #### for DeeperGCN, GINE, GAT, MPNN
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.edge_attr, batch.batch) )
else: #### for GCN, GIN, GraphSAGE
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.batch) )
loss = loss_fn(pred, batch.y[:,0], batch.idx)
optimizer.zero_grad()
loss.backward()
optimizer.step()
######***evaluation***####
# evaluation on training sets
model.eval()
train_pred_list, train_true_list = [], []
with torch.no_grad():
for step, batch in enumerate(trainloader_eval):
batch = batch.cuda()
if with_edge_features: #### for DeeperGCN, GINE, GAT, MPNN
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.edge_attr, batch.batch) )
else: #### for GCN, GIN, GraphSAGE
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.batch) )
train_pred_list.append(pred.cpu().detach().numpy())
train_true_list.append(batch.y[:,0].cpu().detach().numpy())
train_true = np.concatenate(train_true_list)
train_pred = np.concatenate(train_pred_list)
train_ap = auc_prc_score(train_true, train_pred)
train_log.append(train_ap)
# evaluation on test sets
model.eval()
test_pred_list, test_true_list = [], []
with torch.no_grad():
for step, batch in enumerate(testloader):
batch = batch.cuda()
if with_edge_features: #### for DeeperGCN, GINE, GAT, MPNN
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.edge_attr, batch.batch) )
else: #### for GCN, GIN, GraphSAGE
pred = torch.sigmoid( model(batch.x, batch.edge_index, batch.batch) )
test_pred_list.append(pred.cpu().detach().numpy())
test_true_list.append(batch.y[:,0].cpu().detach().numpy())
test_true = np.concatenate(test_true_list)
test_pred = np.concatenate(test_pred_list)
test_ap = auc_prc_score(test_true, test_pred)
test_log.append(test_ap)
if best_test < test_ap:
best_test = test_ap
model.train()
print("epoch: %s, train_ap: %.4f, test_ap: %.4f, best_test_ap: %.4f, lr: %.4f"%(epoch, train_ap, test_ap, best_test, optimizer.lr ))
Start Training
------------------------------
epoch: 0, train_ap: 0.4517, test_ap: 0.3509, best_test_ap: 0.3509, lr: 0.0010
epoch: 1, train_ap: 0.4449, test_ap: 0.3546, best_test_ap: 0.3546, lr: 0.0010
epoch: 2, train_ap: 0.4442, test_ap: 0.3466, best_test_ap: 0.3546, lr: 0.0010
epoch: 3, train_ap: 0.4362, test_ap: 0.3510, best_test_ap: 0.3546, lr: 0.0010
epoch: 4, train_ap: 0.4495, test_ap: 0.3515, best_test_ap: 0.3546, lr: 0.0010
epoch: 5, train_ap: 0.4527, test_ap: 0.3613, best_test_ap: 0.3613, lr: 0.0010
epoch: 6, train_ap: 0.4581, test_ap: 0.3566, best_test_ap: 0.3613, lr: 0.0010
epoch: 7, train_ap: 0.4514, test_ap: 0.3581, best_test_ap: 0.3613, lr: 0.0010
epoch: 8, train_ap: 0.4555, test_ap: 0.3559, best_test_ap: 0.3613, lr: 0.0010
epoch: 9, train_ap: 0.4515, test_ap: 0.3535, best_test_ap: 0.3613, lr: 0.0010
epoch: 10, train_ap: 0.4632, test_ap: 0.3620, best_test_ap: 0.3620, lr: 0.0010
epoch: 11, train_ap: 0.4659, test_ap: 0.3693, best_test_ap: 0.3693, lr: 0.0010
epoch: 12, train_ap: 0.4714, test_ap: 0.3690, best_test_ap: 0.3693, lr: 0.0010
epoch: 13, train_ap: 0.4714, test_ap: 0.3640, best_test_ap: 0.3693, lr: 0.0010
epoch: 14, train_ap: 0.4632, test_ap: 0.3595, best_test_ap: 0.3693, lr: 0.0010
epoch: 15, train_ap: 0.4735, test_ap: 0.3610, best_test_ap: 0.3693, lr: 0.0010
epoch: 16, train_ap: 0.4722, test_ap: 0.3728, best_test_ap: 0.3728, lr: 0.0010
epoch: 17, train_ap: 0.4737, test_ap: 0.3605, best_test_ap: 0.3728, lr: 0.0010
epoch: 18, train_ap: 0.4831, test_ap: 0.3770, best_test_ap: 0.3770, lr: 0.0010
epoch: 19, train_ap: 0.4712, test_ap: 0.3693, best_test_ap: 0.3770, lr: 0.0010
epoch: 20, train_ap: 0.4827, test_ap: 0.3694, best_test_ap: 0.3770, lr: 0.0010
epoch: 21, train_ap: 0.4787, test_ap: 0.3706, best_test_ap: 0.3770, lr: 0.0010
epoch: 22, train_ap: 0.4897, test_ap: 0.3746, best_test_ap: 0.3770, lr: 0.0010
epoch: 23, train_ap: 0.4914, test_ap: 0.3872, best_test_ap: 0.3872, lr: 0.0010
epoch: 24, train_ap: 0.4894, test_ap: 0.3801, best_test_ap: 0.3872, lr: 0.0010
epoch: 25, train_ap: 0.4863, test_ap: 0.3829, best_test_ap: 0.3872, lr: 0.0010
epoch: 26, train_ap: 0.4862, test_ap: 0.3735, best_test_ap: 0.3872, lr: 0.0010
epoch: 27, train_ap: 0.4900, test_ap: 0.3707, best_test_ap: 0.3872, lr: 0.0010
epoch: 28, train_ap: 0.4929, test_ap: 0.3757, best_test_ap: 0.3872, lr: 0.0010
epoch: 29, train_ap: 0.4912, test_ap: 0.3717, best_test_ap: 0.3872, lr: 0.0010
Reducing learning rate to 0.00010 @ T=102660!
epoch: 30, train_ap: 0.5088, test_ap: 0.3881, best_test_ap: 0.3881, lr: 0.0001
epoch: 31, train_ap: 0.5144, test_ap: 0.3878, best_test_ap: 0.3881, lr: 0.0001
epoch: 32, train_ap: 0.5173, test_ap: 0.3886, best_test_ap: 0.3886, lr: 0.0001
epoch: 33, train_ap: 0.5209, test_ap: 0.3892, best_test_ap: 0.3892, lr: 0.0001
epoch: 34, train_ap: 0.5235, test_ap: 0.3899, best_test_ap: 0.3899, lr: 0.0001
epoch: 35, train_ap: 0.5242, test_ap: 0.3904, best_test_ap: 0.3904, lr: 0.0001
epoch: 36, train_ap: 0.5234, test_ap: 0.3883, best_test_ap: 0.3904, lr: 0.0001
epoch: 37, train_ap: 0.5258, test_ap: 0.3880, best_test_ap: 0.3904, lr: 0.0001
epoch: 38, train_ap: 0.5272, test_ap: 0.3893, best_test_ap: 0.3904, lr: 0.0001
epoch: 39, train_ap: 0.5272, test_ap: 0.3901, best_test_ap: 0.3904, lr: 0.0001
epoch: 40, train_ap: 0.5300, test_ap: 0.3908, best_test_ap: 0.3908, lr: 0.0001
epoch: 41, train_ap: 0.5309, test_ap: 0.3904, best_test_ap: 0.3908, lr: 0.0001
epoch: 42, train_ap: 0.5315, test_ap: 0.3886, best_test_ap: 0.3908, lr: 0.0001
epoch: 43, train_ap: 0.5308, test_ap: 0.3881, best_test_ap: 0.3908, lr: 0.0001
epoch: 44, train_ap: 0.5310, test_ap: 0.3890, best_test_ap: 0.3908, lr: 0.0001
Reducing learning rate to 0.00001 @ T=153990!
epoch: 45, train_ap: 0.5333, test_ap: 0.3903, best_test_ap: 0.3908, lr: 0.0000
epoch: 46, train_ap: 0.5337, test_ap: 0.3909, best_test_ap: 0.3909, lr: 0.0000
epoch: 47, train_ap: 0.5339, test_ap: 0.3912, best_test_ap: 0.3912, lr: 0.0000
epoch: 48, train_ap: 0.5341, test_ap: 0.3911, best_test_ap: 0.3912, lr: 0.0000
epoch: 49, train_ap: 0.5344, test_ap: 0.3913, best_test_ap: 0.3913, lr: 0.0000
epoch: 50, train_ap: 0.5350, test_ap: 0.3915, best_test_ap: 0.3915, lr: 0.0000
epoch: 51, train_ap: 0.5347, test_ap: 0.3906, best_test_ap: 0.3915, lr: 0.0000
epoch: 52, train_ap: 0.5358, test_ap: 0.3910, best_test_ap: 0.3915, lr: 0.0000
epoch: 53, train_ap: 0.5352, test_ap: 0.3902, best_test_ap: 0.3915, lr: 0.0000
epoch: 54, train_ap: 0.5344, test_ap: 0.3901, best_test_ap: 0.3915, lr: 0.0000
epoch: 55, train_ap: 0.5355, test_ap: 0.3909, best_test_ap: 0.3915, lr: 0.0000
epoch: 56, train_ap: 0.5353, test_ap: 0.3901, best_test_ap: 0.3915, lr: 0.0000
epoch: 57, train_ap: 0.5365, test_ap: 0.3906, best_test_ap: 0.3915, lr: 0.0000
epoch: 58, train_ap: 0.5362, test_ap: 0.3906, best_test_ap: 0.3915, lr: 0.0000
epoch: 59, train_ap: 0.5367, test_ap: 0.3916, best_test_ap: 0.3916, lr: 0.0000
Visualization
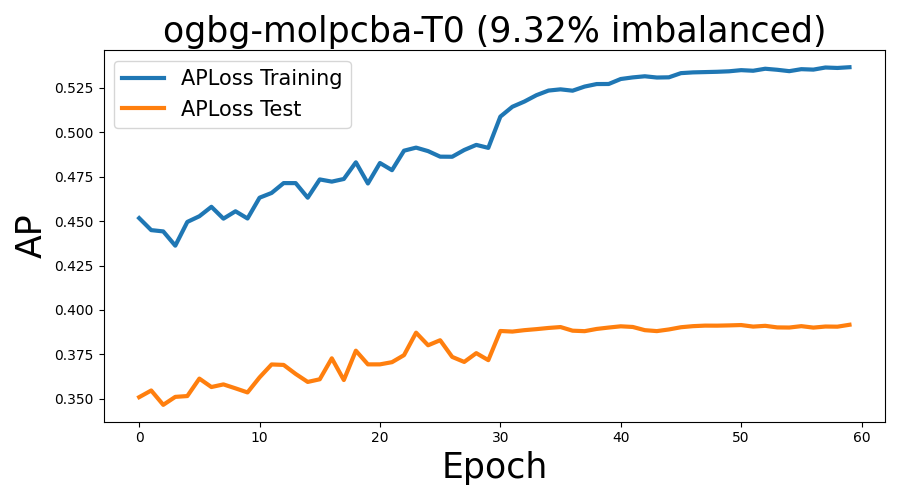
Now, let’s see the learning curve for optimizing AUPRC on train and test sets.
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (9,5)
x=np.arange(len(train_log))
plt.figure()
plt.plot(x, train_log, linestyle='-', label='APLoss Training', linewidth=3)
plt.plot(x, test_log, linestyle='-', label='APLoss Test', linewidth=3)
plt.title('ogbg-molpcba-T0 (9.32% imbalanced)',fontsize=25)
plt.legend(fontsize=15)
plt.ylabel('AP', fontsize=25)
plt.xlabel('Epoch', fontsize=25)
plt.tight_layout()
plt.show()