Multiple Instance Deep AUC Maximization with attention pooling (MIDAM-att) on Histopathology (Image) Dataset
Introduction
In this tutorial, we will learn how to quickly train a ResNet20 model by optimizing Multiple Instance Deep AUC Maximization (MIDAM) under our novel MIDAMLoss(mode='attention') and MIDAM optimizer [Ref] method on a binary classification task on Breast Cancer Histopathology dataset. After completion of this tutorial, you should be able to use LibAUC to train your own models on your own datasets.
Reference:
If you find this tutorial helpful in your work, please cite our library paper and the following papers:
@inproceedings{zhu2023mil,
title={Provable Multi-instance Deep AUC Maximization with Stochastic Pooling},
author={Zhu, Dixian and Wang, Bokun and Chen, Zhi and Wang, Yaxing and Sonka, Milan and Wu, Xiaodong and Yang, Tianbao},
booktitle={International Conference on Machine Learning},
year={2023},
organization={PMLR}
}
Install LibAUC
Let’s start with installing our library here. In this tutorial, we will use the lastest version for LibAUC by using pip install -U.
!pip install -U libauc
Importing LibAUC
Import required libraries to use
import torch
import matplotlib.pyplot as plt
import numpy as np
from libauc.optimizers import MIDAM
from libauc.losses import MIDAMLoss
from libauc.models import ResNet20_stoc_att
from libauc.utils import set_all_seeds, collate_fn, MIL_sampling, MIL_evaluate_auc
from libauc.sampler import DualSampler
from libauc.datasets import BreastCancer, CustomDataset
Reproducibility
These functions limit the number of sources of randomness behaviors, such as model intialization, data shuffling, etcs. However, completely reproducible results are not guaranteed across PyTorch releases [Ref].
def set_all_seeds(SEED):
# REPRODUCIBILITY
torch.manual_seed(SEED)
np.random.seed(SEED)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
Introduction for Loss and Optimizer
In this section, we will introduce pAUC optimization algorithm and how to utilize MIDAMLoss function and MIDAM optimizer.
HyperParameters
The hyper-parameters: batch size (bag-level), instance batch size (instance-level), postive sampling rate, learning rate, weight decay and margin for AUC loss.
# HyperParameters
SEED = 123
set_all_seeds(SEED)
batch_size = 8
instance_batch_size = 128
sampling_rate = 0.5
lr = 5e-2
weight_decay = 5e-4
margin = 0.1
momentum = 0.1
gamma = 0.9
Load Data, initialize model and loss
In this step, we will use the Breast Cancer as benchmark dataset [Ref]. Import data to dataloader. We extend the traditional ResNet20 with an additional attention module: ResNet20_stoc_att. The data are arranged with the shape: (num_bag, num_instance_in_bag, C, H, W)
(train_data, train_labels), (test_data, test_labels) = BreastCancer(MIL_flag=True)
traindSet = CustomDataset(train_data, train_labels, return_index=True)
testSet = CustomDataset(test_data, test_labels, return_index=True)
DIMS=166
sampler = DualSampler(dataset=traindSet, batch_size=batch_size, shuffle=True, sampling_rate=sampling_rate)
trainloader = torch.utils.data.DataLoader(dataset=traindSet, sampler=sampler, batch_size=batch_size, shuffle=False, collate_fn=None)
testloader = torch.utils.data.DataLoader(testSet, batch_size=batch_size, shuffle=False, collate_fn=None)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = ResNet20_stoc_att(num_classes=1).to(device)
Loss = MIDAMLoss(mode='attention',data_len=len(traindSet), gamma=gamma, margin=margin)
optimizer = MIDAM(model.parameters(), loss_fn=Loss, lr=lr, weight_decay=weight_decay, momentum=momentum)
The data shapes for training data/label and testing data/label:
Downloading https://objects.githubusercontent.com/github-production-release-asset-2e65be/647580747/d75046eb-60ac-47e1-b732-67cc3c71e49a?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIWNJYAX4CSVEH53A%2F20230609%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20230609T164941Z&X-Amz-Expires=300&X-Amz-Signature=3089bd6659de76da484b10a9c75dfd83be65ee4dcbe4af6c7dd672b167d2e6bc&X-Amz-SignedHeaders=host&actor_id=0&key_id=0&repo_id=647580747&response-content-disposition=attachment%3B%20filename%3Dbreast.npz&response-content-type=application%2Foctet-stream to ./data/Breast_Cancer/breast.npz 100%|██████████| 239469738/239469738 [00:03<00:00, 72018432.08it/s] (52, 672, 3, 32, 32) (52, 1) (6, 672, 3, 32, 32) (6, 1)
Training
total_epochs = 100
decay_epoch = [50, 75]
train_auc = np.zeros(total_epochs)
test_auc = np.zeros(total_epochs)
for epoch in range(total_epochs):
if epoch in decay_epoch:
optimizer.update_lr(decay_factor=10)
Loss.update_smoothing(decay_factor=2)
for idx, data in enumerate(trainloader):
y_pred = []
sd = []
train_data_bags, train_labels, ids = data
for i in range(len(ids)):
tmp_pred, tmp_sd = MIL_sampling(bag_X=train_data_bags[i], model=model, instance_batch_size=instance_batch_size, mode='att')
y_pred.append(tmp_pred)
sd.append(tmp_sd)
y_pred = torch.cat(y_pred, dim=0)
sd = torch.cat(sd, dim=0)
ids = torch.from_numpy(np.array(ids))
train_labels = torch.from_numpy(np.array(train_labels))
loss = Loss(y_pred=(y_pred,sd), y_true=train_labels, index=ids)
optimizer.zero_grad()
loss.backward()
optimizer.step()
model.eval()
with torch.no_grad():
single_tr_auc = MIL_evaluate_auc(trainloader, model, mode='att')
single_te_auc = MIL_evaluate_auc(testloader, model, mode='att')
train_auc[epoch] = single_tr_auc
test_auc[epoch] = single_te_auc
model.train()
print ('Epoch=%s, BatchID=%s, Tr_AUC=%.4f, Test_AUC=%.4f, lr=%.4f'%(epoch, idx, single_tr_auc, single_te_auc, optimizer.lr))
Epoch=0, BatchID=6, Tr_AUC=0.7258, Test_AUC=0.8750, lr=0.0500
Epoch=1, BatchID=6, Tr_AUC=0.6760, Test_AUC=0.8750, lr=0.0500
Epoch=2, BatchID=6, Tr_AUC=0.7321, Test_AUC=1.0000, lr=0.0500
Epoch=3, BatchID=6, Tr_AUC=0.7054, Test_AUC=0.8750, lr=0.0500
Epoch=4, BatchID=6, Tr_AUC=0.6480, Test_AUC=0.8750, lr=0.0500
Epoch=5, BatchID=6, Tr_AUC=0.7194, Test_AUC=1.0000, lr=0.0500
Epoch=6, BatchID=6, Tr_AUC=0.6837, Test_AUC=1.0000, lr=0.0500
Epoch=7, BatchID=6, Tr_AUC=0.6862, Test_AUC=1.0000, lr=0.0500
Epoch=8, BatchID=6, Tr_AUC=0.7462, Test_AUC=1.0000, lr=0.0500
Epoch=9, BatchID=6, Tr_AUC=0.7015, Test_AUC=1.0000, lr=0.0500
Epoch=10, BatchID=6, Tr_AUC=0.7360, Test_AUC=1.0000, lr=0.0500
Epoch=11, BatchID=6, Tr_AUC=0.7679, Test_AUC=1.0000, lr=0.0500
Epoch=12, BatchID=6, Tr_AUC=0.7717, Test_AUC=1.0000, lr=0.0500
Epoch=13, BatchID=6, Tr_AUC=0.7538, Test_AUC=1.0000, lr=0.0500
Epoch=14, BatchID=6, Tr_AUC=0.7449, Test_AUC=1.0000, lr=0.0500
Epoch=15, BatchID=6, Tr_AUC=0.7168, Test_AUC=1.0000, lr=0.0500
Epoch=16, BatchID=6, Tr_AUC=0.6505, Test_AUC=1.0000, lr=0.0500
Epoch=17, BatchID=6, Tr_AUC=0.7653, Test_AUC=1.0000, lr=0.0500
Epoch=18, BatchID=6, Tr_AUC=0.7921, Test_AUC=1.0000, lr=0.0500
Epoch=19, BatchID=6, Tr_AUC=0.7819, Test_AUC=1.0000, lr=0.0500
Epoch=20, BatchID=6, Tr_AUC=0.7411, Test_AUC=1.0000, lr=0.0500
Epoch=21, BatchID=6, Tr_AUC=0.7870, Test_AUC=1.0000, lr=0.0500
Epoch=22, BatchID=6, Tr_AUC=0.7385, Test_AUC=1.0000, lr=0.0500
Epoch=23, BatchID=6, Tr_AUC=0.7436, Test_AUC=1.0000, lr=0.0500
Epoch=24, BatchID=6, Tr_AUC=0.8202, Test_AUC=1.0000, lr=0.0500
Epoch=25, BatchID=6, Tr_AUC=0.8061, Test_AUC=1.0000, lr=0.0500
Epoch=26, BatchID=6, Tr_AUC=0.8036, Test_AUC=1.0000, lr=0.0500
Epoch=27, BatchID=6, Tr_AUC=0.8010, Test_AUC=1.0000, lr=0.0500
Epoch=28, BatchID=6, Tr_AUC=0.8253, Test_AUC=1.0000, lr=0.0500
Epoch=29, BatchID=6, Tr_AUC=0.8010, Test_AUC=1.0000, lr=0.0500
Epoch=30, BatchID=6, Tr_AUC=0.7436, Test_AUC=1.0000, lr=0.0500
Epoch=31, BatchID=6, Tr_AUC=0.8112, Test_AUC=1.0000, lr=0.0500
Epoch=32, BatchID=6, Tr_AUC=0.8163, Test_AUC=1.0000, lr=0.0500
Epoch=33, BatchID=6, Tr_AUC=0.8099, Test_AUC=1.0000, lr=0.0500
Epoch=34, BatchID=6, Tr_AUC=0.8342, Test_AUC=1.0000, lr=0.0500
Epoch=35, BatchID=6, Tr_AUC=0.8087, Test_AUC=1.0000, lr=0.0500
Epoch=36, BatchID=6, Tr_AUC=0.8087, Test_AUC=1.0000, lr=0.0500
Epoch=37, BatchID=6, Tr_AUC=0.8176, Test_AUC=1.0000, lr=0.0500
Epoch=38, BatchID=6, Tr_AUC=0.8227, Test_AUC=1.0000, lr=0.0500
Epoch=39, BatchID=6, Tr_AUC=0.8495, Test_AUC=1.0000, lr=0.0500
Epoch=40, BatchID=6, Tr_AUC=0.8508, Test_AUC=1.0000, lr=0.0500
Epoch=41, BatchID=6, Tr_AUC=0.8431, Test_AUC=1.0000, lr=0.0500
Epoch=42, BatchID=6, Tr_AUC=0.8036, Test_AUC=1.0000, lr=0.0500
Epoch=43, BatchID=6, Tr_AUC=0.8546, Test_AUC=1.0000, lr=0.0500
Epoch=44, BatchID=6, Tr_AUC=0.8584, Test_AUC=1.0000, lr=0.0500
Epoch=45, BatchID=6, Tr_AUC=0.8355, Test_AUC=1.0000, lr=0.0500
Epoch=46, BatchID=6, Tr_AUC=0.8291, Test_AUC=1.0000, lr=0.0500
Epoch=47, BatchID=6, Tr_AUC=0.8482, Test_AUC=1.0000, lr=0.0500
Epoch=48, BatchID=6, Tr_AUC=0.8444, Test_AUC=1.0000, lr=0.0500
Epoch=49, BatchID=6, Tr_AUC=0.8508, Test_AUC=1.0000, lr=0.0500
Reducing learning rate to 0.00500 @ T=350!
Updating regularizer @ T=350!
Epoch=50, BatchID=6, Tr_AUC=0.8495, Test_AUC=1.0000, lr=0.0050
Epoch=51, BatchID=6, Tr_AUC=0.8304, Test_AUC=1.0000, lr=0.0050
Epoch=52, BatchID=6, Tr_AUC=0.8635, Test_AUC=1.0000, lr=0.0050
Epoch=53, BatchID=6, Tr_AUC=0.8776, Test_AUC=1.0000, lr=0.0050
Epoch=54, BatchID=6, Tr_AUC=0.8418, Test_AUC=1.0000, lr=0.0050
Epoch=55, BatchID=6, Tr_AUC=0.8240, Test_AUC=1.0000, lr=0.0050
Epoch=56, BatchID=6, Tr_AUC=0.8622, Test_AUC=1.0000, lr=0.0050
Epoch=57, BatchID=6, Tr_AUC=0.8635, Test_AUC=1.0000, lr=0.0050
Epoch=58, BatchID=6, Tr_AUC=0.8559, Test_AUC=1.0000, lr=0.0050
Epoch=59, BatchID=6, Tr_AUC=0.8508, Test_AUC=1.0000, lr=0.0050
Epoch=60, BatchID=6, Tr_AUC=0.8291, Test_AUC=1.0000, lr=0.0050
Epoch=61, BatchID=6, Tr_AUC=0.8240, Test_AUC=1.0000, lr=0.0050
Epoch=62, BatchID=6, Tr_AUC=0.8661, Test_AUC=1.0000, lr=0.0050
Epoch=63, BatchID=6, Tr_AUC=0.8559, Test_AUC=1.0000, lr=0.0050
Epoch=64, BatchID=6, Tr_AUC=0.8304, Test_AUC=1.0000, lr=0.0050
Epoch=65, BatchID=6, Tr_AUC=0.8444, Test_AUC=1.0000, lr=0.0050
Epoch=66, BatchID=6, Tr_AUC=0.8482, Test_AUC=1.0000, lr=0.0050
Epoch=67, BatchID=6, Tr_AUC=0.8801, Test_AUC=1.0000, lr=0.0050
Epoch=68, BatchID=6, Tr_AUC=0.8355, Test_AUC=1.0000, lr=0.0050
Epoch=69, BatchID=6, Tr_AUC=0.8699, Test_AUC=1.0000, lr=0.0050
Epoch=70, BatchID=6, Tr_AUC=0.8367, Test_AUC=1.0000, lr=0.0050
Epoch=71, BatchID=6, Tr_AUC=0.8482, Test_AUC=1.0000, lr=0.0050
Epoch=72, BatchID=6, Tr_AUC=0.8597, Test_AUC=1.0000, lr=0.0050
Epoch=73, BatchID=6, Tr_AUC=0.8673, Test_AUC=1.0000, lr=0.0050
Epoch=74, BatchID=6, Tr_AUC=0.8673, Test_AUC=1.0000, lr=0.0050
Reducing learning rate to 0.00050 @ T=525!
Updating regularizer @ T=525!
Epoch=75, BatchID=6, Tr_AUC=0.8367, Test_AUC=1.0000, lr=0.0005
Epoch=76, BatchID=6, Tr_AUC=0.9005, Test_AUC=1.0000, lr=0.0005
Epoch=77, BatchID=6, Tr_AUC=0.8686, Test_AUC=1.0000, lr=0.0005
Epoch=78, BatchID=6, Tr_AUC=0.8661, Test_AUC=1.0000, lr=0.0005
Epoch=79, BatchID=6, Tr_AUC=0.8750, Test_AUC=1.0000, lr=0.0005
Epoch=80, BatchID=6, Tr_AUC=0.8508, Test_AUC=1.0000, lr=0.0005
Epoch=81, BatchID=6, Tr_AUC=0.8597, Test_AUC=1.0000, lr=0.0005
Epoch=82, BatchID=6, Tr_AUC=0.8520, Test_AUC=1.0000, lr=0.0005
Epoch=83, BatchID=6, Tr_AUC=0.8214, Test_AUC=1.0000, lr=0.0005
Epoch=84, BatchID=6, Tr_AUC=0.8061, Test_AUC=1.0000, lr=0.0005
Epoch=85, BatchID=6, Tr_AUC=0.8737, Test_AUC=1.0000, lr=0.0005
Epoch=86, BatchID=6, Tr_AUC=0.8686, Test_AUC=1.0000, lr=0.0005
Epoch=87, BatchID=6, Tr_AUC=0.8469, Test_AUC=1.0000, lr=0.0005
Epoch=88, BatchID=6, Tr_AUC=0.8635, Test_AUC=1.0000, lr=0.0005
Epoch=89, BatchID=6, Tr_AUC=0.8686, Test_AUC=1.0000, lr=0.0005
Epoch=90, BatchID=6, Tr_AUC=0.8431, Test_AUC=1.0000, lr=0.0005
Epoch=91, BatchID=6, Tr_AUC=0.9031, Test_AUC=1.0000, lr=0.0005
Epoch=92, BatchID=6, Tr_AUC=0.8673, Test_AUC=1.0000, lr=0.0005
Epoch=93, BatchID=6, Tr_AUC=0.8495, Test_AUC=1.0000, lr=0.0005
Epoch=94, BatchID=6, Tr_AUC=0.8712, Test_AUC=1.0000, lr=0.0005
Epoch=95, BatchID=6, Tr_AUC=0.8469, Test_AUC=1.0000, lr=0.0005
Epoch=96, BatchID=6, Tr_AUC=0.8571, Test_AUC=1.0000, lr=0.0005
Epoch=97, BatchID=6, Tr_AUC=0.8329, Test_AUC=1.0000, lr=0.0005
Epoch=98, BatchID=6, Tr_AUC=0.8750, Test_AUC=1.0000, lr=0.0005
Epoch=99, BatchID=6, Tr_AUC=0.8648, Test_AUC=1.0000, lr=0.0005
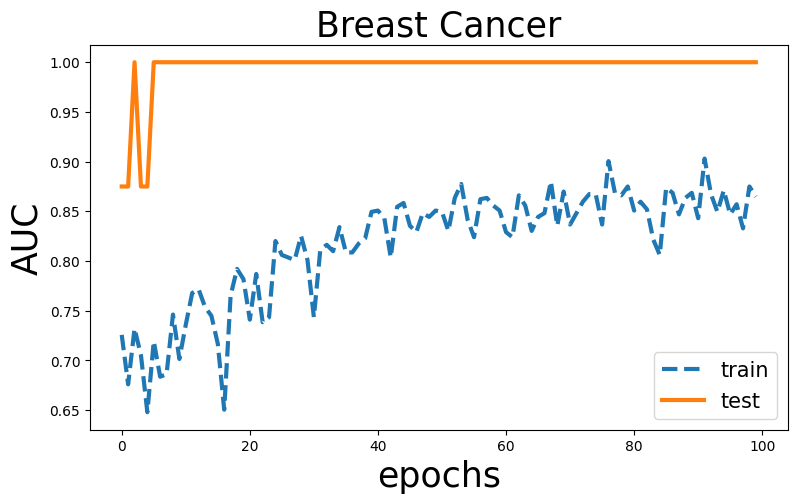
Visualization
plt.rcParams["figure.figsize"] = (9,5)
x=np.arange(len(train_auc))
plt.figure()
plt.plot(x, train_auc, linestyle='--', label='train', linewidth=3)
plt.plot(x, test_auc, label='test', linewidth=3)
plt.title('Breast Cancer',fontsize=25)
plt.legend(fontsize=15)
plt.ylabel('AUC',fontsize=25)
plt.xlabel('epochs',fontsize=25)
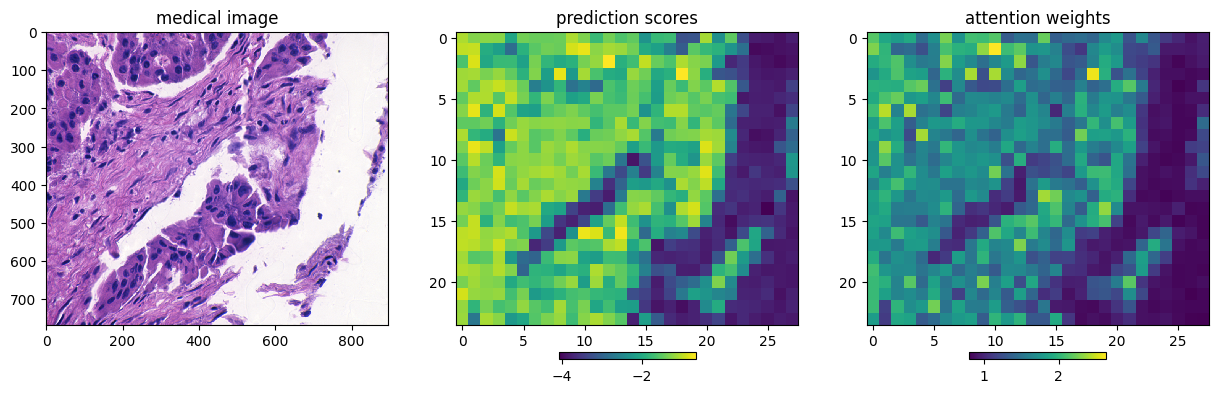
Ablation Study on Attention Weights
maxV = -1e10
maxbag = 0
for idx, data in enumerate(trainloader):
y_pred = []
sd = []
train_data_bags, train_labels, ids = data
for i in range(len(ids)):
y_pred_bag, weights_bag = model(train_data_bags[i].float().cuda())
value = torch.sum(y_pred_bag*weights_bag)/torch.sum(weights_bag)
if value > maxV:
maxV = value
maxbag = train_data_bags[i]
maxlabel = train_labels[i]
print(maxV)
print(maxlabel)
tensor(-1.9086, device='cuda:0', grad_fn=<DivBackward0>)
tensor([1.], dtype=torch.float64)
from mpl_toolkits.axes_grid1 import make_axes_locatable
tmp = maxbag
tmp = tmp.numpy()
tmp = np.transpose(tmp,[0,2,3,1])
img = []
for i in range(24): # collate patches back to the original image arrangement
tmpimg = []
for j in range(28):
tmpimg.append(tmp[i*28+j])
tmpimg = np.concatenate(tmpimg, axis=1)
img.append(tmpimg)
img = np.concatenate(img, axis=0)
y_pred_bag, weights_bag = model(maxbag.float().cuda())
weights = weights_bag.detach().cpu().numpy()
weights = np.reshape(weights, [24,28])
preds = y_pred_bag.detach().cpu().numpy()
preds = np.reshape(preds, [24,28])
fig, subfigs = plt.subplots(figsize=(15, 6), ncols=3)
imgax = subfigs[0]
predax = subfigs[1]
attax = subfigs[2]
imgfig = imgax.imshow(img, interpolation='nearest')
imgax.set_title('medical image')
predfig = predax.imshow(preds, interpolation=None, norm=None)
predax.set_title('prediction scores')
attfig = attax.imshow(weights, interpolation=None, norm=None)
attax.set_title('attention weights')
divider = make_axes_locatable(imgax)
cax = divider.new_vertical(size='5%', pad=0.1, pack_start = True)
plt.colorbar(imgfig, cax=cax, shrink=0.0, orientation = 'horizontal', pad=0.06)
plt.colorbar(predfig, ax=predax, shrink=0.4, orientation = 'horizontal', pad=0.06)
plt.colorbar(attfig, ax=attax, shrink=0.4, orientation = 'horizontal', pad=0.06)
plt.show()